Package Development in R
Fabio Votta
favstats
@favstats
www.favstats.eu
develop-rpkgs.netlify.app
2020-11-04
It's normal to struggle at first but it gets better!
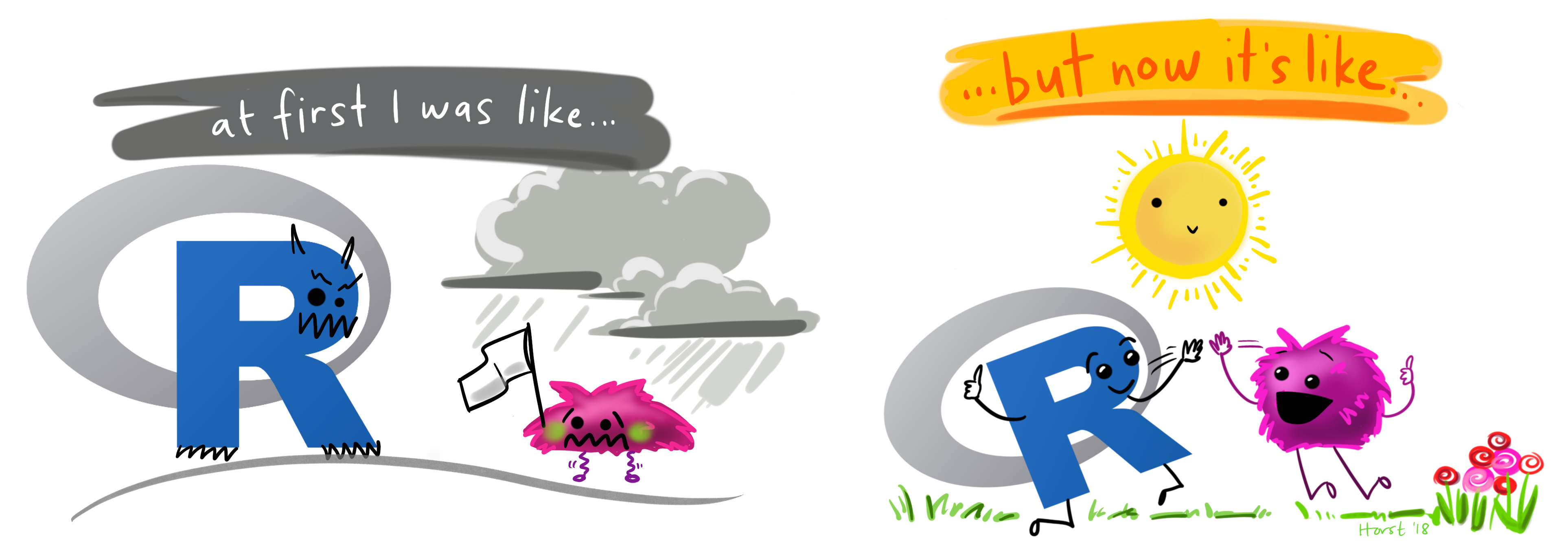
Illustration adapted from Allison Horst
- My experience is that this stuff isn't super easy... but it gets better!
It's normal to struggle at first but it gets better!

Illustration adapted from Allison Horst
My experience is that this stuff isn't super easy... but it gets better!
Mostly because of:
- Awesome inclusive community that is always ready to help
- Great documentation of existing packages and functions
- Active blogosphere with use cases and examples
- and much more!
Overview
Introduction
usethismagicCreating a Package
Documentation (
roxygen2& somedevtools)Unit Tests (with
testthat)Some Resources
Why Even Create an R Package
- R packages are a wonderful way to make functions and datasets easily accessible for everyone
Why Even Create an R Package
R packages are a wonderful way to make functions and datasets easily accessible for everyone
R really comes alive because of the many awesome packages that hundreds and hundreds of volunteers help create and maintain
Why Even Create an R Package
R packages are a wonderful way to make functions and datasets easily accessible for everyone
R really comes alive because of the many awesome packages that hundreds and hundreds of volunteers help create and maintain
It's a great way to become active in the R community & give back
Why Even Create an R Package
R packages are a wonderful way to make functions and datasets easily accessible for everyone
R really comes alive because of the many awesome packages that hundreds and hundreds of volunteers help create and maintain
It's a great way to become active in the R community & give back

Before we Start..
- Creating packages is a process
Before we Start..
Creating packages is a process
There's no need to do everything at once!
Before we Start..
Creating packages is a process
There's no need to do everything at once!
Come up with milestones and focus on reaching them
Before we Start..
Creating packages is a process
There's no need to do everything at once!
Come up with milestones and focus on reaching them
Ask yourself: what's the main purpose of your package?
- Should your package be about ...
- Data Visualization
- Implemetiation of statistical models
- Wrapping an existing API
- Data wrangling
- ???
- a little bit of everything?
Before we Start..
Make sure you installed the latest R and Rstudio version
Install the following packages:
pkgs <- c("devtools", "roxygen2", "usethis", "testthat")install.packages(pkgs)Before we Start..
Make sure you installed the latest R and Rstudio version
Install the following packages:
pkgs <- c("devtools", "roxygen2", "usethis", "testthat")install.packages(pkgs)And now?
Before we Start..
Make sure you installed the latest R and Rstudio version
Install the following packages:
pkgs <- c("devtools", "roxygen2", "usethis", "testthat")install.packages(pkgs)And now?
Choosing a package name

Choosing a Package Name
The available package will help check whether your desired package name is still.. well.. available:
library(available)available("datenguideR", browse = F)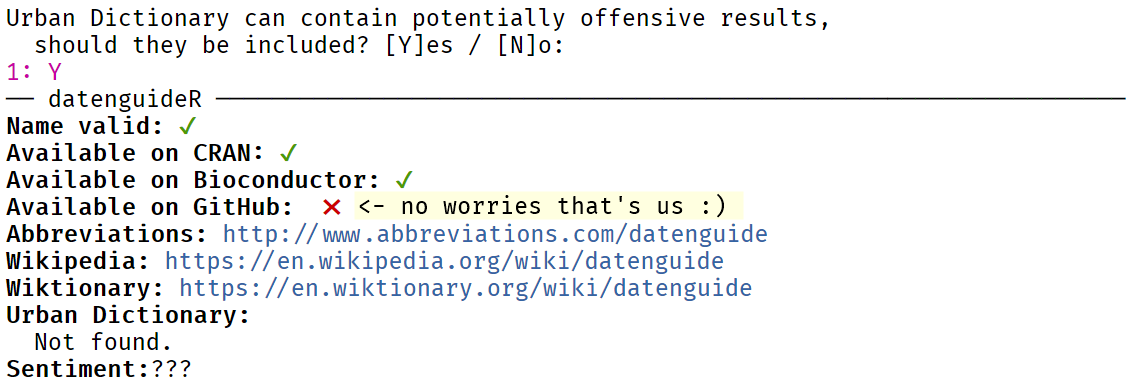
tfw your chosen package name is still available

Now we're ready to create our first R package!
Now we're ready to create our first R package!
With the help of
usethis
usethis
The purpose of usethis is to

… automate repetitive tasks that arise during project setup and development, both for R packages and non-package projects.
usethis
The purpose of usethis is to

… automate repetitive tasks that arise during project setup and development, both for R packages and non-package projects.
As you can guess by the description usethis is VERY useful for package creation and we will be using its functions quite often during development.
usethis
The purpose of usethis is to

… automate repetitive tasks that arise during project setup and development, both for R packages and non-package projects.
As you can guess by the description usethis is VERY useful for package creation and we will be using its functions quite often during development.
We're going to focus on two types of functions within usethis
(although there are MANY more)
usethis
The purpose of usethis is to

… automate repetitive tasks that arise during project setup and development, both for R packages and non-package projects.
As you can guess by the description usethis is VERY useful for package creation and we will be using its functions quite often during development.
We're going to focus on two types of functions within usethis
(although there are MANY more)
use_*- for example
use_pipeto include the pipe operator in your package
- for example
create_*- for example
create_from_githubwhich creates a local Git repository from GitHub
- for example
Creating your first R Package
The following code will create a minimal R package:

library(usethis)create_package("~/git_projects/datenguideR")All you need to do is specify a path. If it exists, it is used. If it does not exist, it is created, provided that the parent path exists.
Creating your first R Package
The following code will create a minimal R package:

library(usethis)create_package("~/git_projects/datenguideR")All you need to do is specify a path. If it exists, it is used. If it does not exist, it is created, provided that the parent path exists.
But because being tidy is awesome we will be using create_tidy_package
create_tidy_package("~/git_projects/datenguideR")This function will also create a new package but it will also apply many great tidyverse conventions that will come in handy.
Creating your first R Package
These files should be in your working directory after creating a tidy package:
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdCreating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis is a folder which holds .R files with your package functions.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis is a folder which holds tests for your package functions. We will talk about tests in greater detail later.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis is a file that contains meta-info about your package including authors, description and license. It's also the place for adding package dependencies.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis file holds namespaces of the package and is auto-generated by ´roxygen2´ (more later). So we don't have to worry about it at all.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdUse this file to communicate with CRAN when you submit a package to their repository. The text should provide an overview of how your package performs on different operating systems.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis file holds some template text for the license under which you publish your package.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis is an Rmarkdown file that can be used to generate the README of your package. The knitted version will be displayed on GitHub.
Creating your first R Package
-- R-- tests-- DESCRIPTION-- NAMESPACE-- cran-comments.md-- LICENSE.md-- README.Rmd-- README.mdThis is a markdown file that was generated by the README.Rmd file. You don't have to worry about this file.
Creating your first R Package
Finally, the following two lines of code will set up Git and a GitHub repository for your package:
use_git()use_github()This is important because people need to be able to install your package from a public source. GitHub is primary source for that but there are others (Gitlab for example).
Creating your first R Package
Finally, the following two lines of code will set up Git and a GitHub repository for your package:
use_git()use_github()This is important because people need to be able to install your package from a public source. GitHub is primary source for that but there are others (Gitlab for example).
Once you have run the two code pieces above you can do the following to install an already fully functioning R package on your computer.
devtools::install_github("{github_username}/{packagename}")Congratulations you created your very first R package!


Let's start adding functions to our package!

Let's start adding functions to our package!
By first of all talking about how to name our functions
Naming Functions
The rOpenSci Package guide states that:
Functions and arguments naming should be chosen to work together to form a common, logical programming API that is easy to read, and auto-complete.
Naming Functions
The rOpenSci Package guide states that:
Functions and arguments naming should be chosen to work together to form a common, logical programming API that is easy to read, and auto-complete.
So in the best case scenario you come up with a naming scheme in the following style:
object_verb
This scheme
- helps avoid namespace conflicts with packages that may have similar verbs
- makes code readable and easy to auto-complete
Naming Functions
The rOpenSci Package guide states that:
Functions and arguments naming should be chosen to work together to form a common, logical programming API that is easy to read, and auto-complete.
So in the best case scenario you come up with a naming scheme in the following style:
object_verb
This scheme
- helps avoid namespace conflicts with packages that may have similar verbs
- makes code readable and easy to auto-complete
For example stringr functions all start with str_* whereas memer functions all start with meme_*.
In our examples here we won't be using this naming style just for convenience but in your personal package you should absolutely consider using it!
It's time to type some R code

Creating your first Function in your Package
In order to get started with our first function we will create a new R script with the help of the usethis package:
use_r("hello")This will create and open an hello.R file in your package's R subfolder; the place where all the functions of your package will live!
Creating your first Function in your Package
In order to get started with our first function we will create a new R script with the help of the usethis package:
use_r("hello")This will create and open an hello.R file in your package's R subfolder; the place where all the functions of your package will live!
Let's create an easy function that will add two numbers:
add <- function(x, y) { x + y}add(1, 2)## [1] 3So far so good.
But how do we communicate what functions do
to people who use our package?
Documentation
With the help of D O C U M E N T A T I O N, of course!
always ⊂_ヽ \\ document \( ͡° ͜ʖ ͡°) > ⌒ヽ / へ\ / / \\your レ ノ ヽ_つ / / / /| ( (ヽ | |、\functions | 丿 \ ⌒) | | ) / ノ ) Lノ (_/Documentation
Documentation is absolutely central.

It's basically the guidance for your users so they know how to use the functions within your package.
Documentation
Documentation is absolutely central.

It's basically the guidance for your users so they know how to use the functions within your package.
Or as Hadley Wickham puts it:
Documentation is one of the most important aspects of good code. Without it, users won’t know how to use your package, and are unlikely to do so.
Documentation
Documentation is absolutely central.

It's basically the guidance for your users so they know how to use the functions within your package.
Or as Hadley Wickham puts it:
Documentation is one of the most important aspects of good code. Without it, users won’t know how to use your package, and are unlikely to do so.
There is a built-in functionality within R to document packages: .Rd files that are stored in the man subdirectory of your package. These files use a syntax that is similar to Latex.
Here to help us with documentation is roxygen2 which conveniently creates a lot of the necessary files with its own more intuitive syntax style.
Documentation

This is how documentation for our add function might look like:
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}Documentation

This is how documentation for our add function might look like:
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}Oi. This looks.. different. So what is actually going on here?
Let's take a look at what this piece of roxygen code will generate.
Documentation


Ah. This looks more familiar!
Let's go through the original roxygen code line by line to understand it better.
Documentation

The first line of code highlighted here shows the title.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}This is a (very) short description of your function.
Documentation

You might have noticed this odd code in front of the documentation: #'
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}Roxygen comments start with this symbol in order to distinguish them from your usual comments (#)
Documentation

Next up is the @param name descriptions for our functions.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}These lines of code describe the function’s inputs or parameters. The description usually documents what the parameter does, what the default inputs are (if any) and what the object type should ideally be (e.g., string, numeric vector etc.).
Documentation

We continue with the @return name description.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of \code{x} and \code{y}.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}The @return tag simply describes the output from the function.
By using \code{} we can also make sure that the variables x and y are written in code font.
Documentation

We continue with the @return name description.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of `x` and `y`.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}The @return tag simply describes the output from the function.
However, since we used create_tidy_package we also automatically turned on markdown style syntax within our roxygen descriptions so this style of syntax is not actually necessary here.
Documentation

Next up is the @export tag.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of `x` and `y`.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}This one is pretty straightforward. If you want your functions to be available to the user (and not just create an internal function to be used within the package) then it is crucial to add the @export tag.
Documentation

Finally, the @examples tag.
#' Add together two numbers.#'#' @param x A number.#' @param y A number.#' @return The sum of `x` and `y`.#' @export#' @examples#' add(1, 1)#' add(10, 1)add <- function(x, y) { x + y}Adding examples to functions can really help your users figure out how your package is meant to be used.
Documentation

With this, the roxygen code for your documentation is done. The last thing to do is converting your description to .Rd files so it can appear in the help file.
Luckily, with the help of the devtools package this step is really easy to do.
Just type in the following code:
library(devtools)document()Updating datenguideR documentationWriting NAMESPACELoading datenguideRWriting NAMESPACEWriting add.RdIf you now type ?add into R it should render the development description that we saw earlier.
That's it!
We created our very first function documentation!

However...
this is not everything of course.
There are so much more things to know about documentation!
I recommend the following sources:
Object documentation chapter from R Packages book
Next up:
Just testthat
Unit Testing

<⌒/ヽ-、_ /<_/____/ 'I cant sleep if you dont test your functions' ∧_∧ ( ・ω・) _| ⊃/(___ / └-(____/  ̄ ̄ ̄ ̄ ̄ ̄ ̄Testing is really important to make sure your package is functioning as intended.
Unit Testing

<⌒/ヽ-、_ /<_/____/ 'I cant sleep if you dont test your functions' ∧_∧ ( ・ω・) _| ⊃/(___ / └-(____/  ̄ ̄ ̄ ̄ ̄ ̄ ̄Testing is really important to make sure your package is functioning as intended.
As you keep developing and adding to your package code, keeping track of what might go wrong will become more and more complex.
Unit Testing

<⌒/ヽ-、_ /<_/____/ 'I cant sleep if you dont test your functions' ∧_∧ ( ・ω・) _| ⊃/(___ / └-(____/  ̄ ̄ ̄ ̄ ̄ ̄ ̄Testing is really important to make sure your package is functioning as intended.
As you keep developing and adding to your package code, keeping track of what might go wrong will become more and more complex.
Unit tests will help you with identifying issues within your code so you can pinpoint what went wrong and where.
Unit Testing

The basic setup of an unit testing is quite simple:
You write up example code with your package functions and define specific outputs that you expect.
- If they are met, great, the test is passed!
- If not, the test fails and you will know that something is wrong with your code and what failed.
Unit Testing

The basic setup of an unit testing is quite simple:
You write up example code with your package functions and define specific outputs that you expect.
- If they are met, great, the test is passed!
- If not, the test fails and you will know that something is wrong with your code and what failed.
To set up unit tests for your package with testthat you can simply run use_testthat (from our beloved usethis package):
use_testthat()Unit Testing

The basic setup of an unit testing is quite simple:
You write up example code with your package functions and define specific outputs that you expect.
- If they are met, great, the test is passed!
- If not, the test fails and you will know that something is wrong with your code and what failed.
To set up unit tests for your package with testthat you can simply run use_testthat (from our beloved usethis package):
use_testthat()This will create a folder tests and a subfolder testthat. This is the folder where your tests will live.
Since we created our package with create_tidy_package we don't need to run this part as this function automatically sets up testthat.
Unit Testing

How do set up individual tests?
usethis got us covered again!
Simply run this code:
use_test("hello")Unit Testing

How do set up individual tests?
usethis got us covered again!
Simply run this code:
use_test("hello")This will create the following file:
tests/testthat/test-hello.RUnit Testing

How do set up individual tests?
usethis got us covered again!
Simply run this code:
use_test("hello")This will create the following file:
tests/testthat/test-hello.RWith this content:
test_that("multiplication works", { expect_equal(2 * 2, 4)})Unit Testing

Let's take this short code apart:
test_that("multiplication works", { expect_equal(2 * 2, 4)})In the first line of code we can see the function test_that which is our main tool for unit tests.
Unit Testing

Let's take this short code apart:
test_that("multiplication works", { expect_equal(2 * 2, 4)})In the first line of code we can see the function test_that which is our main tool for unit tests.
The first argument of test_that describes what is supposed to be tested whereas the second argument encapsulates the code to be tested within a curly bracket {} environment.
Unit Testing

Let's take this short code apart:
test_that("multiplication works", { expect_equal(2 * 2, 4)})In the first line of code we can see the function test_that which is our main tool for unit tests.
The first argument of test_that describes what is supposed to be tested whereas the second argument encapsulates the code to be tested within a curly bracket {} environment.
Within this environment we write up our expectations with the help of testthat's expect_* functions, of which there are many.
Unit Testing

Let's take this short code apart:
test_that("multiplication works", { expect_equal(2 * 2, 4)})In the first line of code we can see the function test_that which is our main tool for unit tests.
The first argument of test_that describes what is supposed to be tested whereas the second argument encapsulates the code to be tested within a curly bracket {} environment.
Within this environment we write up our expectations with the help of testthat's expect_* functions, of which there are many.
In this case we use expect_equal which is quite straightforward. The 1st argument should be equal to the 2nd argument and if it's not, the test fails.
You ready?

Good. Because we are now designing our own test for the add function.
Unit Testing

Alright. So let's start testing our add function.
First, we will test whether addition works as intended:
test_that("addition works as intended", { result <- add(5, 5) expect_equal(result, 10)})The test did not throw an error so this worked. Perfect!
Unit Testing

How about testing that supplying a string should throw an error?
test_that("supplying string should throw error", { expect_error( add(5, "i_am_not_a_number") )})Again, this ran without problems. So everything went as we expected!
Unit Testing

Now, let's try to break our tests!

Unit Testing

Let's try to test whether 5 + 5 is more than 15:
test_that("result is more than 15", { result <- add(5, 5) expect_more_than(result, 15)})Unit Testing

Let's try to test whether 5 + 5 is more than 15:
test_that("result is more than 15", { result <- add(5, 5) expect_more_than(result, 15)})Error: Test failed: 'result is more than 15' * `result` is not strictly more than 15. Difference: -5Success! We failed! Since 5 + 5 is not more than 15 (it's actually 5 less) the test fails and we receive an informative error message as well.
Unit Testing

It takes a little bit of creativity to foresee the many ways functions within your package might break.
In practice, writing the right kind of unit tests is considered sort of an art by some and a nuisance by others.
In any case though it is really important for writing robust code that can be relied upon.
Unit Testing
Now, we have succesfully written up three tests for our add function.

The devtools package will help us test all of our unit tests at once by running the following code within your package project:
test()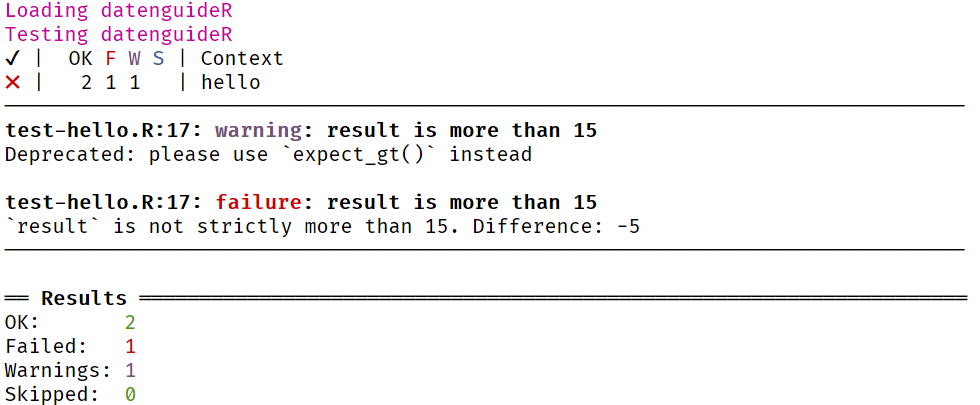
Bringing it all together
We can also run check from devtools to test pretty much everything about our package:

check()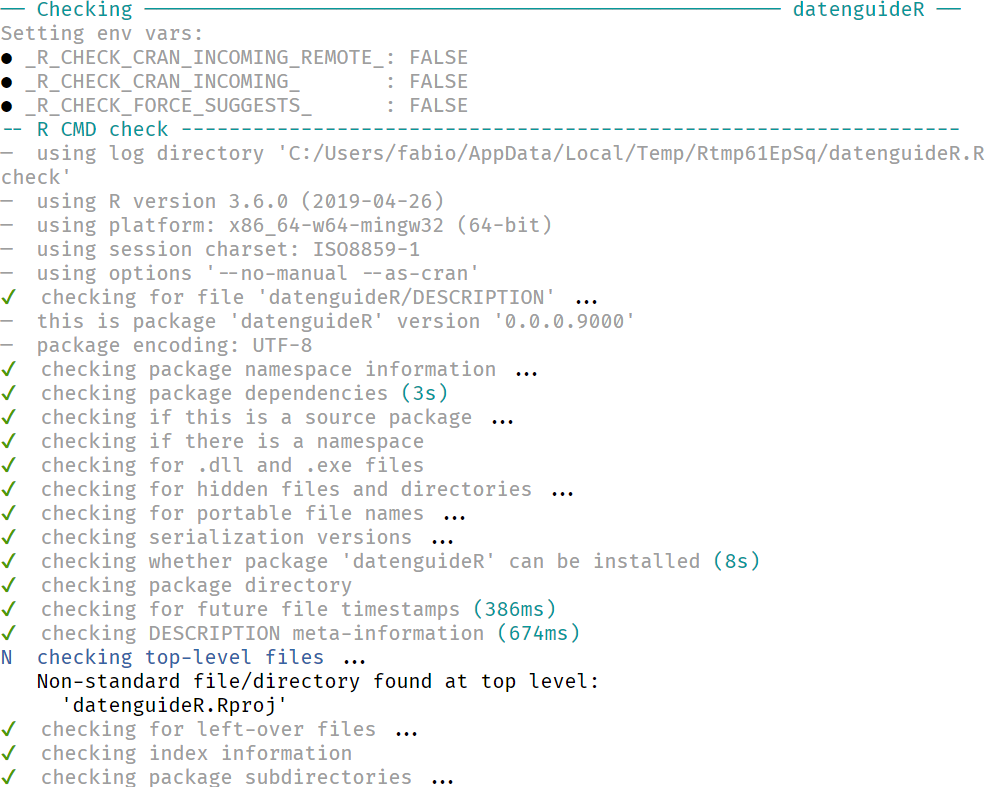
And when your package passes all checks you get the sweet feeling of getting the following feedback:

Useful stuff that I wasn't able to mention yet
Add Package imports to DESCRIPTION with usethis::use_package:
use_package("dplyr")Useful stuff that I wasn't able to mention yet
Add Package imports to DESCRIPTION with usethis::use_package:
use_package("dplyr")Set up continuous integration (in this case Travis CI) with the following code:
use_travis()Continuous integration allows you to run checks on your package code with various R versions and ensures that your package can run on different systems like Linux, Mac OSX & Windows.
You can start reading about Travis CI here: A BEGINNER'S GUIDE TO TRAVIS-CI FOR R.
Useful stuff that I wasn't able to mention yet
Add Package imports to DESCRIPTION with usethis::use_package:
use_package("dplyr")Set up continuous integration (in this case Travis CI) with the following code:
use_travis()Continuous integration allows you to run checks on your package code with various R versions and ensures that your package can run on different systems like Linux, Mac OSX & Windows.
You can start reading about Travis CI here: A BEGINNER'S GUIDE TO TRAVIS-CI FOR R.
Set up an awesome website for your package (documentation) with the help of pkgdown.
use_pkgdown()Thank you for listening
